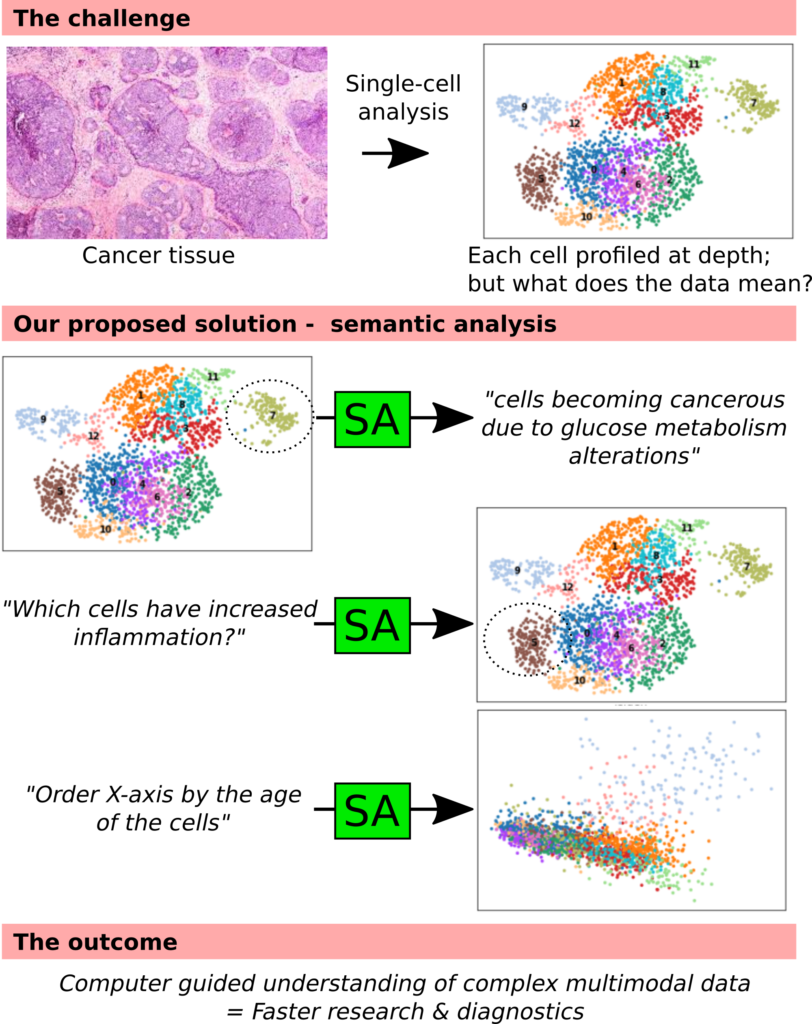
Single-cell analysis is the current state of the art in biology – tissue samples such as cancer can be separated into individual cells, which are then measured at depth using sequencing. Enormous amounts of data are produced, enabling precise investigation of the content of the tissue. The bottleneck is now the researcher, and exploring a single dataset can take up to one year.
In this project we will use text analysis to guide the analysis. Over 500 000 articles have been published about just T cells, and using modern machine learning we will extract the biological knowledge. The researcher will be able to use the system in several ways; (1) asking why cells behave like they do, (2) ask which cells correspond to a certain behavior or (3) organize summaries by descriptions of certain biological phenomena.
The success of this outcome will greatly aid basic research. However, single-cell is increasingly used for advanced diagnostics, and being able to quickly analyze data is of essence for bringing this method to the clinic.
Our approach will use generative large-scale language models, fine-tuned over the available open access literature, curated literature-gene linkages, and other large datasets. The language model will then integrate with a Variational Autoencoder (VAE) model that captures the statistical properties of the single-cell data.
The candidate postdoc should be familiar with transformer models, ideally the GPT family. Furthermore, knowledge of clustering and VAEs is a merit, as is experience of computational linguistics. Willingness to learn basic biological concepts is key to success. The candidate will work in an interdisciplinary team at Umeå University under supervision of Johanna Björklund (department of computing science) and Johan Henriksson (department of molecular biology and MIMS).
If you are interested in this position, please get in touch with either Johan Henriksson or Johanna Björklund.
